Development of the Spatial Multi-Omics Technology MiP-Seq for Deciphering the Spatial Proteome and Transcriptome of the Tumor Microenvironment.
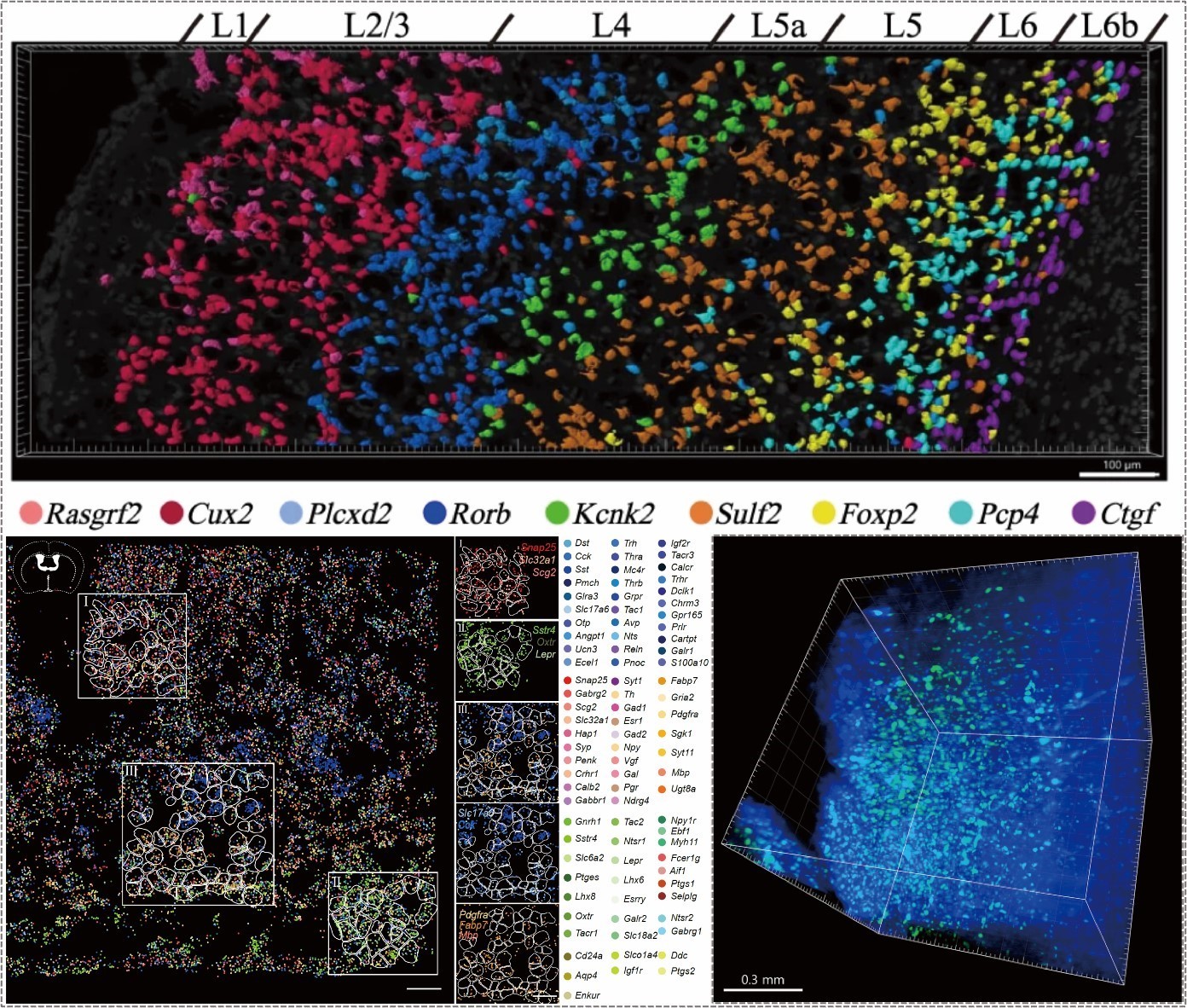
The research team has successfully developed a highly efficient high-throughput paired-end in situ sequencing spatial omics technology. We have established platforms for both in situ spatial omics and targeted spatial omics. By designing padlock probes to directly target specific molecular sequences, we utilize RNA-dependent DNA ligase to form circularized DNA, followed by rolling circle amplification (RCA) to enhance signal detection. Furthermore, we have upgraded the traditional single-barcode encoding system to a dual-barcode combinatorial encoding approach, significantly increasing the detection throughput per sequencing round.